TUTORIAL¶
To be completed...
Basic usage¶
Initialize panel
panel=Panel(fig_width=900,)
Create a track
test_track = tracks.BaseTrack(name = 'test', sort_by = None)
Create a feature
feat1=features.Simple(name = 'feat1', start = 100, end = 356, )
Add a features to a track
test_track.append(feat1)
test_track.extend([feat1, feat2, feat3])
or directly:
test_track = tracks.BaseTrack(features.Simple(name = 'feat1', start = 100, end = 356, ),
features.Simple(name = 'feat2', start = 30, end = 856, ),
features.Simple(name = 'feat3', start = 400, end = 700, ),
sort_by = 'None',)
Add a track to the panel:
panel.add_track(test_track)
Save image. This supports all format supported by matplotlib including:
- png
- jpeg
- svg
panel.save('test_image.png')
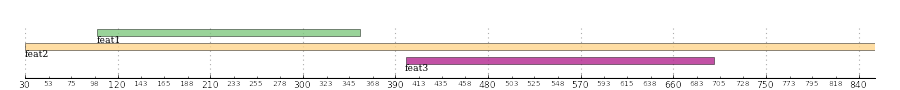
Complete basic example:
from biograpy import Panel, tracks, features
panel=Panel(fig_width=900, padding = 25)#initialize a drawer, and add some padding
test_track = tracks.BaseTrack(features.Simple(name = 'feat1', start = 100, end = 356, ),
features.Simple(name = 'feat2', start = 30, end = 856, ),
features.Simple(name = 'feat3', start = 400, end = 700, ),
sort_by = 'None',)
panel.add_track(test_track)
panel.save('basic_test.png')
And this is the resulting image:
The Panel¶
The panel object is the “canvas” to which all the objects will be drawn.
panel.fig is the matplotlib Figure object
panel.tracks is the collector of all the tracks. It is a Python list object
panel.save(filepath) calling this method will draw all the tracks and features to the panel.fig object, and will save the figure to the filepath passed
panel.htmlmap after panel.save(filepath) has been called the image htmlmap is accessible as this panel property if the specified format was png or jpg.
TODO
Tracks¶
Handling Tracks¶
Tracks can be seen as ordered collection of features.
For convenience track.append() and track.extend works as the common list methods.
TODO
Ordering features in track¶
collapse (default)
order as they come = user defined
order by lenght
order by score
TODO
Features¶
Simple features¶
TODO
Draw complex features¶
mRNA and CDS¶
TODO
Transmembrane regions¶
TODO
Secondary structures¶
TODO
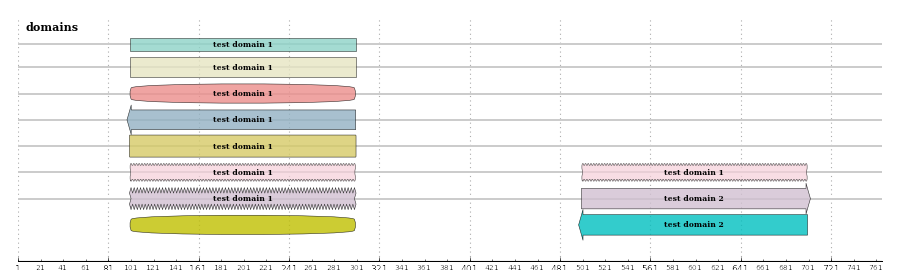
Domains¶
from biograpy import Panel, tracks, features
from Bio import SeqFeature
panel = Panel(fig_width=900)
test_track = tracks.BaseTrack(name = 'domains', sort_by = None, cm = 'Set3')
feat = SeqFeature.SeqFeature()
feat.location = SeqFeature.FeatureLocation(100, 300)
dfeat = features.DomainFeature([feat], name = 'test domain 1', seq_line = (1, 766))
test_track.append(dfeat)
dfeat = features.DomainFeature([feat],name = 'test domain 1', height = 1.5, seq_line = (1, 766))
test_track.append(dfeat)
dfeat = features.DomainFeature([feat],name = 'test domain 1', height = 1.5, boxstyle = 'round4, rounding_size=1.4', seq_line = (1, 766))
test_track.append(dfeat)
dfeat = features.DomainFeature([feat],name = 'test domain 1',height = 1.5, boxstyle = 'larrow, pad = 0', seq_line = (1, 766))
test_track.append(dfeat)
dfeat = features.DomainFeature([feat], name = 'test domain 1',height = 1.5, boxstyle = 'round ', seq_line = (1, 766))
test_track.append(dfeat)
feat2 = SeqFeature.SeqFeature()
feat2.location = SeqFeature.FeatureLocation(500, 700)
dfeat = features.DomainFeature([feat,feat2], name = 'test domain 1',height = 1.5, boxstyle = 'roundtooth, pad = 0.1, tooth_size=1.2', seq_line = (1, 766), alpha = 0.7)
test_track.append(dfeat)
dfeat = features.DomainFeature([feat,feat2],name = ['test domain 1', 'test domain 2'], height = 1.5, boxstyle = ['sawtooth, tooth_size=1.4', 'rarrow, pad = 0.1'], seq_line = (1, 766), ec = 'None')
test_track.append(dfeat)
dfeat = features.DomainFeature([feat,feat2],name = ['', 'test domain 2'],height = 1.5, boxstyle = ['round4, rounding_size=1.4', 'larrow, pad = 0.1'], ec = 'None', fc = ['y', 'c'], color_by_cm = False)
test_track.append(dfeat)
panel.add_track(test_track)
panel.save('domain_test.png')
resulting image:
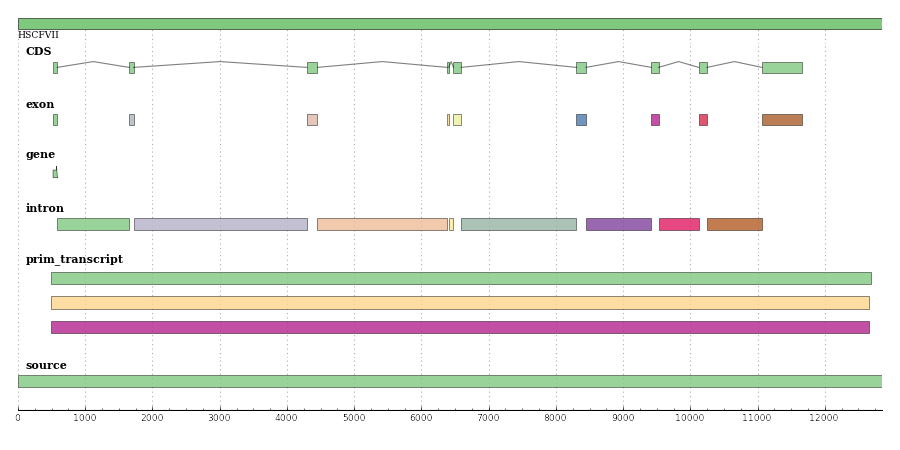
Draw a SeqRecord¶
Starting from this annotated entry in EMBL format
Generate a plot
from biograpy.seqrecord import SeqRecordDrawer
from biograpy import Panel, tracks, features
from Bio import SeqIO
seqrecord = SeqIO.parse(open("factor7.embl"), "embl").next()
SeqRecordDrawer(seqrecord, fig_width=900).save("factor7.png")
Check out the similar plot done with BioPerl here.
Draw a Blast Output¶
from biograpy import Panel, tracks, features
from Bio import SeqIO, AlignIO, SeqFeature
from Bio.Blast import NCBIXML
'''Parse BLAST XML output'''
aligndata = []
blasthits=NCBIXML.parse(open('BLASTalignment.xml'))
id_filter = .3 #show only alignments with sequence identity higher than 30%
for hit in blasthits:
for alignment in hit.alignments:
hsps = []
for hsp in alignment.hsps:
if (hsp.identities/float(hsp.align_length) >= id_filter):
hit_id = alignment.hit_id
hit_name = alignment.hit_def [:40]
align=dict(hit_id = hit_id,
hit_name = hit_name,
hit_start = hsp.sbjct_start,
hit_stop = hsp.sbjct_end,
query_start = hsp.query_start,
query_stop = hsp.query_end,
align_lenght = hsp.align_length,
identity_percent = hsp.identities/float(hsp.align_length)*100,
similarity_percent = hsp.positives/float(hsp.align_length)*100,
evalue = hsp.expect,
blast_score = hsp.score,
query_seq = hsp.query,
match_seq = hsp.match,
hit_seq = hsp.sbjct,
aligns = hsp.num_alignments,
other = dir(hsp))
hsps.append(align)
aligndata.append(dict(hsps= hsps,
hit_def = alignment.hit_def,
hit_id = alignment.hit_id,
length = alignment.length,
title = alignment.title,))
'''Draw Alignment'''
panel=Panel(fig_width=900,)#initialize a drawer
ref_track = tracks.BaseTrack(features.Simple(1,#draw a reference object
hit.query_length,
name = hit.query,
height = 1.5,
fc='grey',
ec='k',),
name = 'Query')
panel.append(ref_track)
aligns_track = tracks.BaseTrack(name = 'Hits', cm ='Blues', )
for align in aligndata:
for hsp in align['hsps']:
location = SeqFeature.FeatureLocation(hsp['query_start'], hsp['query_stop'])
feat = SeqFeature.SeqFeature()
feat.type = 'Hit'
feat.id = hsp['hit_id']
feat.location = location
feat.qualifiers['ft_description'] = ['Hit']
feat.qualifiers['ft_type'] = ['Hit']
feat.qualifiers['score'] = float(hsp['identity_percent'])/100.
feat.qualifiers['hit_name'] = [hsp['hit_id']]
aligns_track.append(features.GenericSeqFeature(feat,
score = feat.qualifiers['score'],
name = hsp['hit_name'],
use_score_for_color = True,
url = "http://www.ncbi.nlm.nih.gov/protein/%s"%hsp['hit_id'].split('|')[-2],))
panel.append(aligns_track)
panel.save('blast_output.png')
print panel.htmlmap
Generated image:

to be coupled with the generated html map available calling
panel.htmlmap
<map name="biograpy-map" id="biograpy-map">
<area shape="rect" coords="18,43,882,28" href="#sp|P04626|ERBB2_HUMAN Receptor tyrosine-protein kinase erbB-2 OS=Homo sapiens GN=ERBB2 PE=1 SV=1" target="_self" alt="sp|P04626|ERBB2_HUMAN Receptor tyrosine-protein kinase erbB-2 OS=Homo sapiens GN=ERBB2 PE=1 SV=1" >
<area shape="rect" coords="18,93,882,81" href="http://www.ncbi.nlm.nih.gov/protein/NP_004439.2" target="_self" alt="receptor tyrosine-protein kinase erbB-2 " >
<area shape="rect" coords="38,116,882,105" href="http://www.ncbi.nlm.nih.gov/protein/NP_001005862.1" target="_self" alt="receptor tyrosine-protein kinase erbB-2 " >
<area shape="rect" coords="33,139,877,128" href="http://www.ncbi.nlm.nih.gov/protein/NP_005219.2" target="_self" alt="epidermal growth factor receptor isoform" >
<area shape="rect" coords="23,163,877,151" href="http://www.ncbi.nlm.nih.gov/protein/NP_001036064.1" target="_self" alt="receptor tyrosine-protein kinase erbB-4 " >
<area shape="rect" coords="32,186,877,174" href="http://www.ncbi.nlm.nih.gov/protein/NP_005226.1" target="_self" alt="receptor tyrosine-protein kinase erbB-4 " >
<area shape="rect" coords="20,209,704,197" href="http://www.ncbi.nlm.nih.gov/protein/NP_001973.2" target="_self" alt="receptor tyrosine-protein kinase erbB-3 " >
<area shape="rect" coords="33,232,452,221" href="http://www.ncbi.nlm.nih.gov/protein/NP_958439.1" target="_self" alt="epidermal growth factor receptor isoform" >
<area shape="rect" coords="33,255,452,244" href="http://www.ncbi.nlm.nih.gov/protein/NP_958441.1" target="_self" alt="epidermal growth factor receptor isoform" >
<area shape="rect" coords="33,279,298,267" href="http://www.ncbi.nlm.nih.gov/protein/NP_958440.1" target="_self" alt="epidermal growth factor receptor isoform" >
<area shape="rect" coords="432,279,706,267" href="http://www.ncbi.nlm.nih.gov/protein/NP_004432.1" target="_self" alt="ephrin type-B receptor 1 precursor [Homo" >
<area shape="rect" coords="479,232,701,221" href="http://www.ncbi.nlm.nih.gov/protein/NP_004422.2" target="_self" alt="ephrin type-A receptor 2 precursor [Homo" >
<area shape="rect" coords="509,255,693,244" href="http://www.ncbi.nlm.nih.gov/protein/NP_001129473.1" target="_self" alt="tyrosine-protein kinase ABL2 isoform e [" >
<area shape="rect" coords="461,302,716,290" href="http://www.ncbi.nlm.nih.gov/protein/NP_872272.2" target="_self" alt="ephrin type-A receptor 5 isoform b precu" >
<area shape="rect" coords="504,325,706,313" href="http://www.ncbi.nlm.nih.gov/protein/NP_004435.3" target="_self" alt="ephrin type-B receptor 4 precursor [Homo" >
<area shape="rect" coords="504,348,716,336" href="http://www.ncbi.nlm.nih.gov/protein/NP_004430.4" target="_self" alt="ephrin type-A receptor 5 isoform a precu" >
<area shape="rect" coords="516,371,693,360" href="http://www.ncbi.nlm.nih.gov/protein/NP_004094.3" target="_self" alt="protein-tyrosine kinase 2-beta isoform a" >
<area shape="rect" coords="516,394,693,383" href="http://www.ncbi.nlm.nih.gov/protein/NP_775267.1" target="_self" alt="protein-tyrosine kinase 2-beta isoform b" >
<area shape="rect" coords="467,418,695,406" href="http://www.ncbi.nlm.nih.gov/protein/NP_004433.2" target="_self" alt="ephrin type-B receptor 2 isoform 2 precu" >
<area shape="rect" coords="467,441,695,429" href="http://www.ncbi.nlm.nih.gov/protein/NP_059145.2" target="_self" alt="ephrin type-B receptor 2 isoform 1 precu" >
<area shape="rect" coords="509,464,693,452" href="http://www.ncbi.nlm.nih.gov/protein/NP_001129472.1" target="_self" alt="tyrosine-protein kinase ABL2 isoform d [" >
<area shape="rect" coords="509,487,693,476" href="http://www.ncbi.nlm.nih.gov/protein/NP_001161711.1" target="_self" alt="tyrosine-protein kinase ABL2 isoform i [" >
<area shape="rect" coords="509,510,693,499" href="http://www.ncbi.nlm.nih.gov/protein/NP_005149.4" target="_self" alt="tyrosine-protein kinase ABL2 isoform c [" >
<area shape="rect" coords="509,534,693,522" href="http://www.ncbi.nlm.nih.gov/protein/NP_001161708.1" target="_self" alt="tyrosine-protein kinase ABL2 isoform f [" >
<area shape="rect" coords="509,557,693,545" href="http://www.ncbi.nlm.nih.gov/protein/NP_009298.1" target="_self" alt="tyrosine-protein kinase ABL2 isoform b [" >
<area shape="rect" coords="509,580,693,568" href="http://www.ncbi.nlm.nih.gov/protein/NP_001161709.1" target="_self" alt="tyrosine-protein kinase ABL2 isoform g [" >
<area shape="rect" coords="509,603,693,592" href="http://www.ncbi.nlm.nih.gov/protein/NP_001161710.1" target="_self" alt="tyrosine-protein kinase ABL2 isoform h [" >
<area shape="rect" coords="503,626,690,615" href="http://www.ncbi.nlm.nih.gov/protein/NP_722560.1" target="_self" alt="focal adhesion kinase 1 isoform a [Homo " >
<area shape="rect" coords="498,650,687,638" href="http://www.ncbi.nlm.nih.gov/protein/NP_005772.3" target="_self" alt="activated CDC42 kinase 1 isoform 1 [Homo" >
<area shape="rect" coords="503,673,695,661" href="http://www.ncbi.nlm.nih.gov/protein/NP_005598.3" target="_self" alt="focal adhesion kinase 1 isoform b [Homo " >
<area shape="rect" coords="517,696,684,684" href="http://www.ncbi.nlm.nih.gov/protein/NP_997402.1" target="_self" alt="tyrosine-protein kinase ZAP-70 isoform 2" >
<area shape="rect" coords="503,719,690,708" href="http://www.ncbi.nlm.nih.gov/protein/NP_001186578.1" target="_self" alt="focal adhesion kinase 1 isoform c [Homo " >
<area shape="rect" coords="498,742,687,731" href="http://www.ncbi.nlm.nih.gov/protein/NP_001010938.1" target="_self" alt="activated CDC42 kinase 1 isoform 2 [Homo" >
<area shape="rect" coords="478,766,706,754" href="http://www.ncbi.nlm.nih.gov/protein/NP_004429.1" target="_self" alt="ephrin type-A receptor 4 precursor [Homo" >
<area shape="rect" coords="504,789,711,777" href="http://www.ncbi.nlm.nih.gov/protein/NP_005224.2" target="_self" alt="ephrin type-A receptor 3 isoform a precu" >
<area shape="rect" coords="502,812,693,800" href="http://www.ncbi.nlm.nih.gov/protein/NP_005148.2" target="_self" alt="tyrosine-protein kinase ABL1 isoform a [" >
<area shape="rect" coords="502,835,693,823" href="http://www.ncbi.nlm.nih.gov/protein/NP_009297.2" target="_self" alt="tyrosine-protein kinase ABL1 isoform b [" >
<area shape="rect" coords="504,858,720,847" href="http://www.ncbi.nlm.nih.gov/protein/NP_004434.2" target="_self" alt="ephrin type-B receptor 3 precursor [Homo" >
<area shape="rect" coords="516,881,724,870" href="http://www.ncbi.nlm.nih.gov/protein/NP_065681.1" target="_self" alt="proto-oncogene tyrosine-protein kinase r" >
<area shape="rect" coords="516,905,724,893" href="http://www.ncbi.nlm.nih.gov/protein/NP_066124.1" target="_self" alt="proto-oncogene tyrosine-protein kinase r" >
<area shape="rect" coords="517,928,684,916" href="http://www.ncbi.nlm.nih.gov/protein/NP_001070.2" target="_self" alt="tyrosine-protein kinase ZAP-70 isoform 1" >
<area shape="rect" coords="516,951,684,939" href="http://www.ncbi.nlm.nih.gov/protein/NP_001128524.1" target="_self" alt="tyrosine-protein kinase SYK isoform 2 [H" >
<area shape="rect" coords="516,974,684,963" href="http://www.ncbi.nlm.nih.gov/protein/NP_003168.2" target="_self" alt="tyrosine-protein kinase SYK isoform 1 [H" >
<area shape="rect" coords="516,997,691,986" href="http://www.ncbi.nlm.nih.gov/protein/NP_000236.2" target="_self" alt="hepatocyte growth factor receptor isofor" >
<area shape="rect" coords="474,1021,706,1009" href="http://www.ncbi.nlm.nih.gov/protein/NP_004431.1" target="_self" alt="ephrin type-A receptor 7 precursor [Homo" >
<area shape="rect" coords="516,1044,691,1032" href="http://www.ncbi.nlm.nih.gov/protein/NP_001120972.1" target="_self" alt="hepatocyte growth factor receptor isofor" >
<area shape="rect" coords="516,1067,692,1055" href="http://www.ncbi.nlm.nih.gov/protein/NP_002438.2" target="_self" alt="macrophage-stimulating protein receptor " >
<area shape="rect" coords="504,1090,701,1079" href="http://www.ncbi.nlm.nih.gov/protein/NP_005223.4" target="_self" alt="ephrin type-A receptor 1 precursor [Homo" >
<area shape="rect" coords="504,1113,695,1102" href="http://www.ncbi.nlm.nih.gov/protein/NP_065387.1" target="_self" alt="ephrin type-A receptor 8 isoform 1 precu" >
<area shape="rect" coords="509,1137,692,1125" href="http://www.ncbi.nlm.nih.gov/protein/NP_005537.3" target="_self" alt="tyrosine-protein kinase ITK/TSK [Homo sa" >
<area shape="rect" coords="512,1160,692,1148" href="http://www.ncbi.nlm.nih.gov/protein/NP_003319.2" target="_self" alt="tyrosine-protein kinase TXK [Homo sapien" >
<area shape="rect" coords="509,1183,691,1171" href="http://www.ncbi.nlm.nih.gov/protein/NP_075254.1" target="_self" alt="fibroblast growth factor receptor 3 isof" >
<area shape="rect" coords="509,1206,691,1195" href="http://www.ncbi.nlm.nih.gov/protein/NP_000133.1" target="_self" alt="fibroblast growth factor receptor 3 isof" >
<area shape="rect" coords="509,1229,691,1218" href="http://www.ncbi.nlm.nih.gov/protein/NP_001156685.1" target="_self" alt="fibroblast growth factor receptor 3 isof" >
<area shape="rect" coords="512,1252,687,1241" href="http://www.ncbi.nlm.nih.gov/protein/NP_003206.2" target="_self" alt="tyrosine-protein kinase Tec [Homo sapien" >
<area shape="rect" coords="516,1276,706,1264" href="http://www.ncbi.nlm.nih.gov/protein/NP_075594.1" target="_self" alt="basic fibroblast growth factor receptor " >
<area shape="rect" coords="516,1299,706,1287" href="http://www.ncbi.nlm.nih.gov/protein/NP_075593.1" target="_self" alt="basic fibroblast growth factor receptor " >
<area shape="rect" coords="509,1322,687,1310" href="http://www.ncbi.nlm.nih.gov/protein/NP_002022.1" target="_self" alt="tyrosine-protein kinase FRK [Homo sapien" >
<area shape="rect" coords="516,1345,692,1334" href="http://www.ncbi.nlm.nih.gov/protein/NP_001167535.1" target="_self" alt="basic fibroblast growth factor receptor " >
<area shape="rect" coords="516,1368,692,1357" href="http://www.ncbi.nlm.nih.gov/protein/NP_001167534.1" target="_self" alt="basic fibroblast growth factor receptor " >
<area shape="rect" coords="516,1392,692,1380" href="http://www.ncbi.nlm.nih.gov/protein/NP_056934.2" target="_self" alt="basic fibroblast growth factor receptor " >
<area shape="rect" coords="516,1415,691,1403" href="http://www.ncbi.nlm.nih.gov/protein/NP_001138391.1" target="_self" alt="fibroblast growth factor receptor 2 isof" >
<area shape="rect" coords="516,1438,692,1426" href="http://www.ncbi.nlm.nih.gov/protein/NP_075598.2" target="_self" alt="basic fibroblast growth factor receptor " >
<area shape="rect" coords="516,1461,691,1450" href="http://www.ncbi.nlm.nih.gov/protein/NP_001138386.1" target="_self" alt="fibroblast growth factor receptor 2 isof" >
<area shape="rect" coords="516,1484,691,1473" href="http://www.ncbi.nlm.nih.gov/protein/NP_001138389.1" target="_self" alt="fibroblast growth factor receptor 2 isof" >
<area shape="rect" coords="516,1508,706,1496" href="http://www.ncbi.nlm.nih.gov/protein/NP_001167538.1" target="_self" alt="basic fibroblast growth factor receptor " >
<area shape="rect" coords="516,1531,691,1519" href="http://www.ncbi.nlm.nih.gov/protein/NP_001138387.1" target="_self" alt="fibroblast growth factor receptor 2 isof" >
<area shape="rect" coords="516,1554,691,1542" href="http://www.ncbi.nlm.nih.gov/protein/NP_001138390.1" target="_self" alt="fibroblast growth factor receptor 2 isof" >
<area shape="rect" coords="516,1577,691,1566" href="http://www.ncbi.nlm.nih.gov/protein/NP_001138388.1" target="_self" alt="fibroblast growth factor receptor 2 isof" >
<area shape="rect" coords="513,1600,695,1589" href="http://www.ncbi.nlm.nih.gov/protein/NP_002028.1" target="_self" alt="tyrosine-protein kinase Fyn isoform a [H" >
<area shape="rect" coords="516,1624,691,1612" href="http://www.ncbi.nlm.nih.gov/protein/NP_075259.4" target="_self" alt="fibroblast growth factor receptor 2 isof" >
<area shape="rect" coords="516,1647,691,1635" href="http://www.ncbi.nlm.nih.gov/protein/NP_001138385.1" target="_self" alt="fibroblast growth factor receptor 2 isof" >
<area shape="rect" coords="516,1670,691,1658" href="http://www.ncbi.nlm.nih.gov/protein/NP_000132.3" target="_self" alt="fibroblast growth factor receptor 2 isof" >
<area shape="rect" coords="452,1693,691,1682" href="http://www.ncbi.nlm.nih.gov/protein/NP_006334.2" target="_self" alt="tyrosine-protein kinase Mer precursor [H" >
<area shape="rect" coords="480,1716,693,1705" href="http://www.ncbi.nlm.nih.gov/protein/NP_543013.1" target="_self" alt="tyrosine-protein kinase Srms [Homo sapie" >
<area shape="rect" coords="516,1739,695,1728" href="http://www.ncbi.nlm.nih.gov/protein/NP_001229708.1" target="_self" alt="tyrosine-protein kinase Fyn isoform d [H" >
<area shape="rect" coords="512,1763,688,1751" href="http://www.ncbi.nlm.nih.gov/protein/NP_000052.1" target="_self" alt="tyrosine-protein kinase BTK [Homo sapien" >
<area shape="rect" coords="513,1786,688,1774" href="http://www.ncbi.nlm.nih.gov/protein/NP_001706.2" target="_self" alt="tyrosine-protein kinase Blk [Homo sapien" >
<area shape="rect" coords="495,1809,691,1797" href="http://www.ncbi.nlm.nih.gov/protein/NP_004374.1" target="_self" alt="tyrosine-protein kinase CSK [Homo sapien" >
<area shape="rect" coords="516,1832,695,1821" href="http://www.ncbi.nlm.nih.gov/protein/NP_694592.1" target="_self" alt="tyrosine-protein kinase Fyn isoform b [H" >
<area shape="rect" coords="516,1855,705,1844" href="http://www.ncbi.nlm.nih.gov/protein/NP_075252.2" target="_self" alt="fibroblast growth factor receptor 4 isof" >
<area shape="rect" coords="516,1879,705,1867" href="http://www.ncbi.nlm.nih.gov/protein/NP_002002.3" target="_self" alt="fibroblast growth factor receptor 4 isof" >
<area shape="rect" coords="467,1902,687,1890" href="http://www.ncbi.nlm.nih.gov/protein/NP_002935.2" target="_self" alt="proto-oncogene tyrosine-protein kinase R" >
<area shape="rect" coords="513,1925,687,1913" href="http://www.ncbi.nlm.nih.gov/protein/NP_005408.1" target="_self" alt="proto-oncogene tyrosine-protein kinase S" >
<area shape="rect" coords="432,1948,697,1937" href="http://www.ncbi.nlm.nih.gov/protein/NP_000450.2" target="_self" alt="angiopoietin-1 receptor precursor [Homo " >
<area shape="rect" coords="499,1971,687,1960" href="http://www.ncbi.nlm.nih.gov/protein/NP_000206.2" target="_self" alt="tyrosine-protein kinase JAK3 [Homo sapie" >
<area shape="rect" coords="517,1995,688,1983" href="http://www.ncbi.nlm.nih.gov/protein/NP_005239.1" target="_self" alt="tyrosine-protein kinase Fgr [Homo sapien" >
<area shape="rect" coords="495,2018,705,2006" href="http://www.ncbi.nlm.nih.gov/protein/NP_647611.1" target="_self" alt="megakaryocyte-associated tyrosine-protei" >
<area shape="rect" coords="465,2041,703,2029" href="http://www.ncbi.nlm.nih.gov/protein/NP_996844.1" target="_self" alt="leukocyte tyrosine kinase receptor isofo" >
<area shape="rect" coords="465,2064,703,2053" href="http://www.ncbi.nlm.nih.gov/protein/NP_002335.2" target="_self" alt="leukocyte tyrosine kinase receptor isofo" >
<area shape="rect" coords="460,2087,697,2076" href="http://www.ncbi.nlm.nih.gov/protein/NP_055030.1" target="_self" alt="insulin receptor-related protein precurs" >
<area shape="rect" coords="523,2111,695,2099" href="http://www.ncbi.nlm.nih.gov/protein/NP_694593.1" target="_self" alt="tyrosine-protein kinase Fyn isoform c [H" >
<area shape="rect" coords="495,2134,705,2122" href="http://www.ncbi.nlm.nih.gov/protein/NP_002369.2" target="_self" alt="megakaryocyte-associated tyrosine-protei" >
<area shape="rect" coords="495,2157,705,2145" href="http://www.ncbi.nlm.nih.gov/protein/NP_647612.1" target="_self" alt="megakaryocyte-associated tyrosine-protei" >
<area shape="rect" coords="487,2180,691,2169" href="http://www.ncbi.nlm.nih.gov/protein/NP_006284.2" target="_self" alt="tyrosine-protein kinase receptor TYRO3 [" >
<area shape="rect" coords="509,2203,688,2192" href="http://www.ncbi.nlm.nih.gov/protein/NP_001712.1" target="_self" alt="cytoplasmic tyrosine-protein kinase BMX " >
<area shape="rect" coords="465,2226,702,2215" href="http://www.ncbi.nlm.nih.gov/protein/NP_001690.2" target="_self" alt="tyrosine-protein kinase receptor UFO iso" >
<area shape="rect" coords="465,2250,702,2238" href="http://www.ncbi.nlm.nih.gov/protein/NP_068713.2" target="_self" alt="tyrosine-protein kinase receptor UFO iso" >
<area shape="rect" coords="501,2273,709,2261" href="http://www.ncbi.nlm.nih.gov/protein/NP_000199.2" target="_self" alt="insulin receptor isoform Long preproprot" >
<area shape="rect" coords="501,2296,709,2284" href="http://www.ncbi.nlm.nih.gov/protein/NP_001073285.1" target="_self" alt="insulin receptor isoform Short prepropro" >
</map>
or just save as an SVG file to get href attributes automatically added:
panel.save('blast_output.svg')